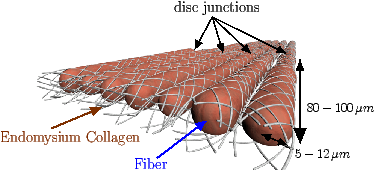
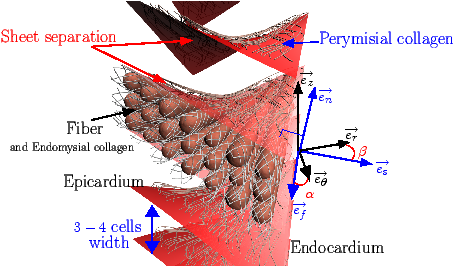
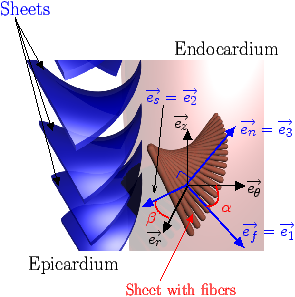
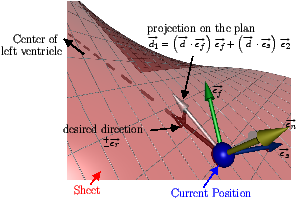
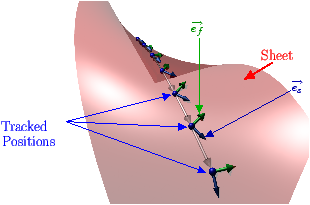
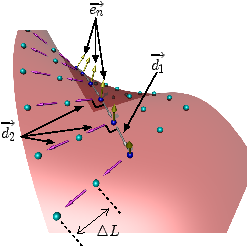
[]
[]
[]

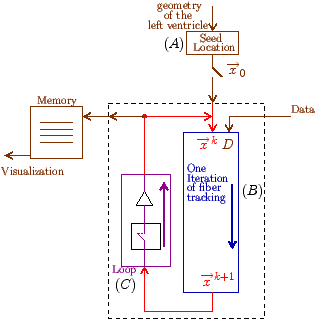
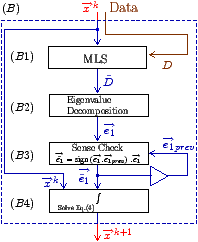
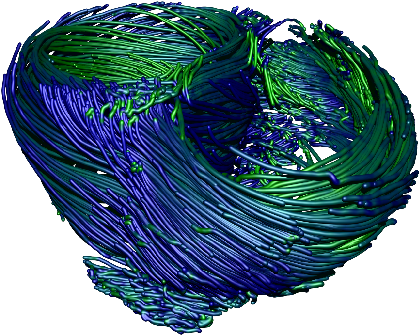
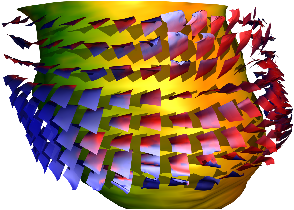
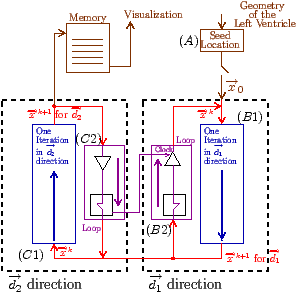
Fig. 7: [
Sheet Tracking Algorithm]Block diagram of the sheet reconstruction algorithm. Picture (a) shows the propagation of the information around the main blocks. Block (A) is the initial position where the sheet tracking starts. Block (B1) corresponds to the iteration in the cross direction and bloc (C1) represents the iteration in the circumferential direction. For each iteration the position on the surface
xk+1 is stored and after triangulation is sent to the visualization module. (B2) and (C2) are the looping blocks with the switch and the buffer to save the previous position. The two loops are synchronized to do all the iterations in the
d2 directions for each in
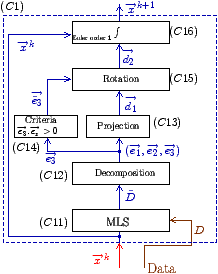
d1. Picture (b) describes the circumferential iteration. (C11) is the MLS filtering and the decomposition is done in (C12). There is the projection in (C13) and a positive sense is taken for
e 3 in (C14) to realize the rotation of
d1 in (C15). Then the integration step is performed in (C16). Picture (c) is the description of the cross section iteration. (B11) is the MLS regularization, (B12) is the decomposition into eigenvectors, the projection to get
d1 is realized in (B13) and the integration step is performed in (B14).